10. Align double ended measurements
The cable length was initially configured during the DTS measurement. For double ended measurements it is important to enter the correct length so that the forward channel and the backward channel are aligned.
This notebook shows how to better align the forward and the backward measurements. Do this before the calibration steps.
[1]:
import os
from dtscalibration import read_silixa_files
from dtscalibration.dts_accessor_utils import (
suggest_cable_shift_double_ended,
shift_double_ended,
)
import numpy as np
%matplotlib inline
[2]:
?suggest_cable_shift_double_ended
[3]:
filepath = os.path.join("..", "..", "tests", "data", "double_ended2")
ds_aligned = read_silixa_files(
directory=filepath, timezone_netcdf="UTC", file_ext="*.xml"
) # this one is already correctly aligned
6 files were found, each representing a single timestep
6 recorded vars were found: LAF, ST, AST, REV-ST, REV-AST, TMP
Recorded at 1693 points along the cable
The measurement is double ended
Reading the data from disk
Because our loaded files were already nicely aligned, we are purposely offsetting the forward and backward channel by 3 `spacial indices’.
[4]:
ds_notaligned = shift_double_ended(ds_aligned, 3)
/home/docs/checkouts/readthedocs.org/user_builds/python-dts-calibration/envs/latest/lib/python3.9/site-packages/dtscalibration/dts_accessor_utils.py:964: UserWarning: The following variables could not be shifted and are not included in the new dataset: ['tmp']. You can silence this warning with the keyword argument `verbose=False`.
warnings.warn(msg, UserWarning)
The device-calibrated temperature doesnot have a valid meaning anymore and is dropped
[5]:
suggested_shift = suggest_cable_shift_double_ended(
ds_notaligned, np.arange(-5, 5), plot_result=True, figsize=(12, 8)
)
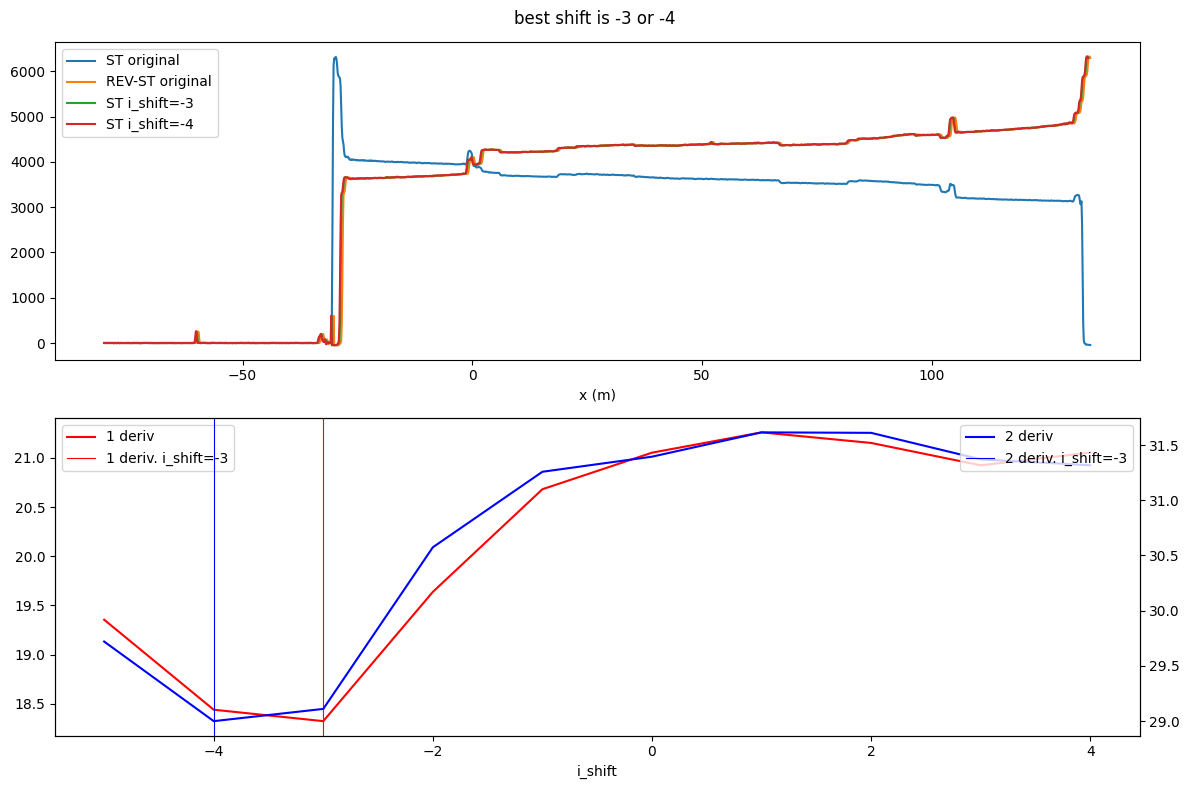
The two approaches suggest a shift of -3 and -4. It is up to the user which suggestion to follow. Usually the two suggested shift are close
[6]:
ds_restored = shift_double_ended(ds_notaligned, suggested_shift[0])
[7]:
print(ds_aligned.x, 3 * "\n", ds_restored.x)
<xarray.DataArray 'x' (x: 1693)> Size: 14kB
array([-80.5043, -80.3772, -80.2501, ..., 134.294 , 134.421 , 134.548 ])
Coordinates:
* x (x) float64 14kB -80.5 -80.38 -80.25 -80.12 ... 134.3 134.4 134.5
Attributes:
name: distance
description: Length along fiber
long_description: Starting at connector of forward channel
units: m
<xarray.DataArray 'x' (x: 1687)> Size: 13kB
array([-80.123 , -79.9959, -79.8688, ..., 133.913 , 134.04 , 134.167 ])
Coordinates:
* x (x) float64 13kB -80.12 -80.0 -79.87 -79.74 ... 133.9 134.0 134.2
Attributes:
name: distance
description: Length along fiber
long_description: Starting at connector of forward channel
units: m
Note that our fiber has become shorter by 2*3 spatial indices